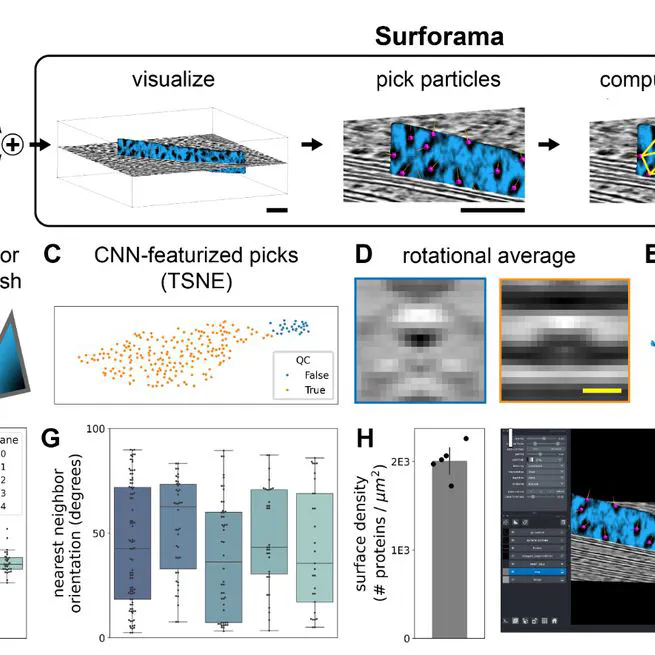
Surforama is a Python package and napari plugin for interactive visualization, annotation, and analysis of proteins in cryo-ET data, with results exportable as RELION-formatted STAR files.
Jul 2, 2024
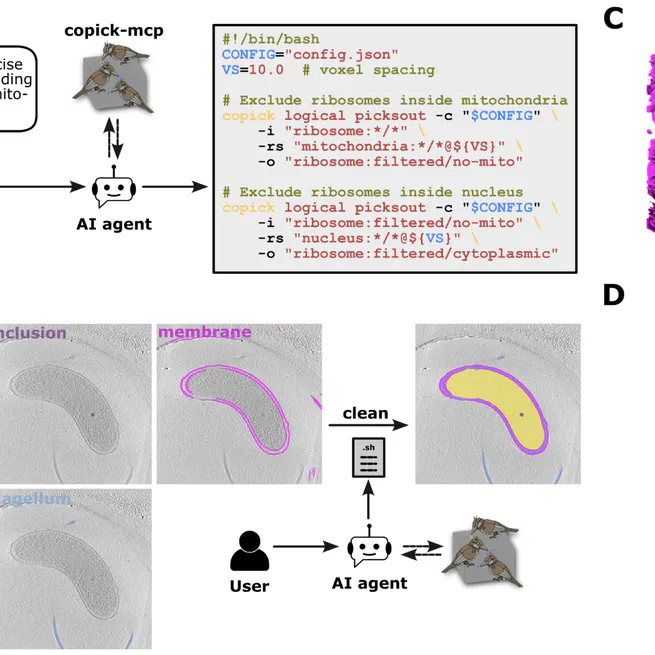
copick is a cross-platform API for collaborative work on cryo-electron tomography datasets. It provides standardized access to tomograms, picks, and segmentations across different storage backends—whether your data is local, on S3, or from the CZ CryoET Data Portal. Built to support the CZII CryoET ML competition on Kaggle, copick enables you to write data handling code once and have it work across diverse storage systems. This makes it easier to share annotations and build ML pipelines that scale across distributed data sources. Code: github.com/copick/copick
Jan 1, 2024
This paper presents napari-imagej, a plugin that integrates the ImageJ ecosystem with napari, enabling access to ImageJ functions directly from napari without the need for extensive porting or maintenance.
Oct 23, 2023
napari is a Python viewer for multi-dimensional images. I joined CZI’s napari team in 2022, where I made code contributions and served as tech lead. I’ve been on napari’s steering council since 2023. napari has become central infrastructure in the scientific Python ecosystem for microscopy—used for everything from basic image visualization to interactive ML annotation workflows. My contributions have focused on large-data handling, ML integration, and helping coordinate the project’s technical direction.
Jul 1, 2022
Album is a software tool for sharing scientific code and software early in the development process. The goal is accelerating the iterative development of scientific software between developers and users. Check out the repository here.
Jul 8, 2020