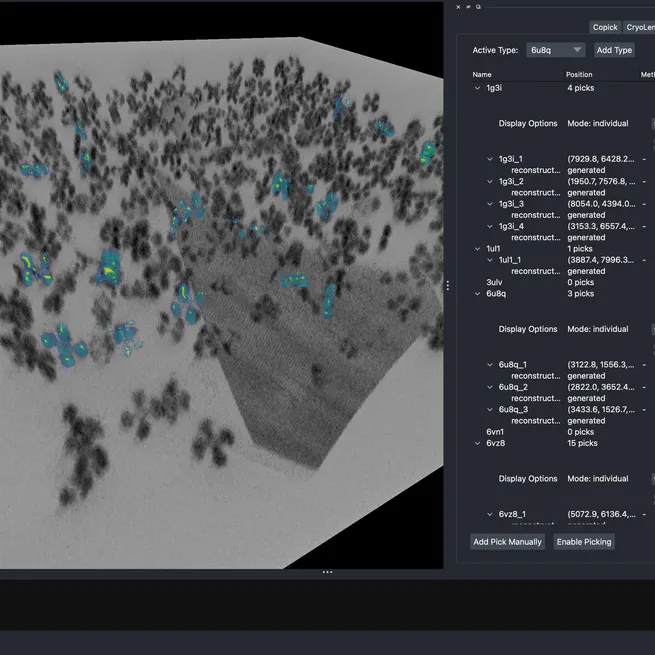
CryoLens is a 3D variational autoencoder for learning interpretable representations of cellular structures from cryo-electron tomography data (available on the CZI Virtual Cell Platform). The model learns compressed representations of 3D subtomograms that capture structural variation in an interpretable latent space—useful for visualization, classification, and understanding structural diversity. Trained on a large synthetic dataset composed of 100+ diverse structures, CryoLens can produce high-fidelity single-angle 3D densities. Tools are available for napari and the Virtual Cell Platform.
Nov 5, 2025
Invited talk on interactive ML for cellular architecture analysis.
Jun 1, 2025
The CZII CryoET Object Identification Challenge, hosted on Kaggle from November 2024 to February 2025, was a machine learning competition focused on automating the detection of protein complexes in cryo-electron tomography (cryoET) data. Published in Nature Methods (Peck et al., 2025), this competition addressed a critical bottleneck in structural biology research by developing algorithms to accurately identify and annotate multiple types of protein complexes in 3D tomographic volumes. More on the Kaggle page
Nov 6, 2024
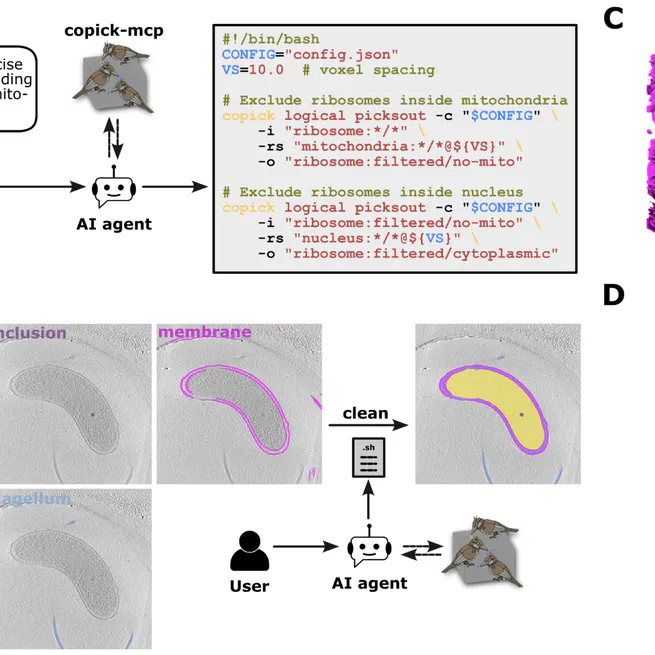
copick is a cross-platform API for collaborative work on cryo-electron tomography datasets. It provides standardized access to tomograms, picks, and segmentations across different storage backends—whether your data is local, on S3, or from the CZ CryoET Data Portal. Built to support the CZII CryoET ML competition on Kaggle, copick enables you to write data handling code once and have it work across diverse storage systems. This makes it easier to share annotations and build ML pipelines that scale across distributed data sources. Code: github.com/copick/copick
Jan 1, 2024