Surforama: interactive exploration of volumetric data by leveraging 3D surfaces
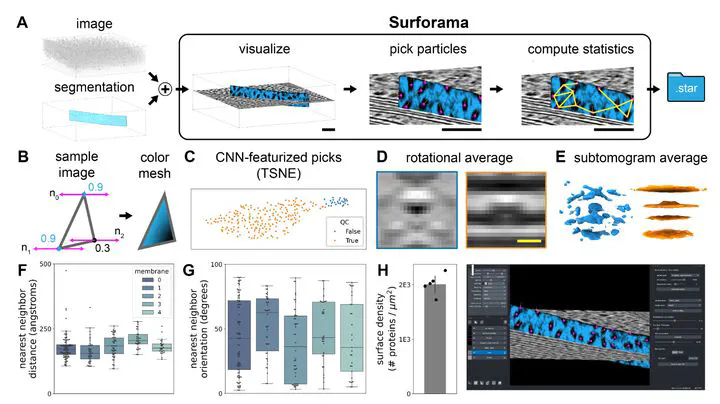 Image credit:
Image credit:Motivation: Visualization and annotation of segmented surfaces is of paramount importance for studying membrane proteins in their native cellular environment by cryogenic electron tomography (cryo-ET). Yet, analyzing membrane proteins and their organization is challenging due to their small sizes and the need to consider local context constrained to the membrane surface.
Results: To interactively visualize, annotate, and analyze proteins in cellular context from cryo-ET data, we have developed Surforama, a Python package and napari plugin. For interactive visualization of membrane proteins in tomograms, Surforama renders the local densities projected on the surface of the segmentations. Surforama additionally provides tools to annotate and analyze particles on the membrane surfaces. Finally, for compatibility with other tools in the cryo-ET analysis ecosystem, results can be exported as RELION-formatted STAR files. As a demonstration, we performed subtomogram averaging and neighborhood analysis of photosystem II proteins in thylakoid membranes from the green alga Chlamydomonas reinhardtii.