A realistic phantom dataset for benchmarking cryo-ET data annotation
Jan 1, 2025·,,,,,,,,,,,,,,,,,,,·
0 min read
Ariana Peck
Yue Yu
Jonathan Schwartz
Anchi Cheng
Utz Heinrich Ermel
Joshua Hutchings
Saugat Kandel
Dari Kimanius
Elizabeth a Montabana
Daniel Serwas
Hannah Siems
Feng Wang
Zhuowen Zhao
Shawn Zheng
Matthias Haury
David Agard
Clinton Potter
Bridget Carragher
Kyle Harrington, * Co-Corresponding
Mohammadreza Paraan, * Co-Corresponding
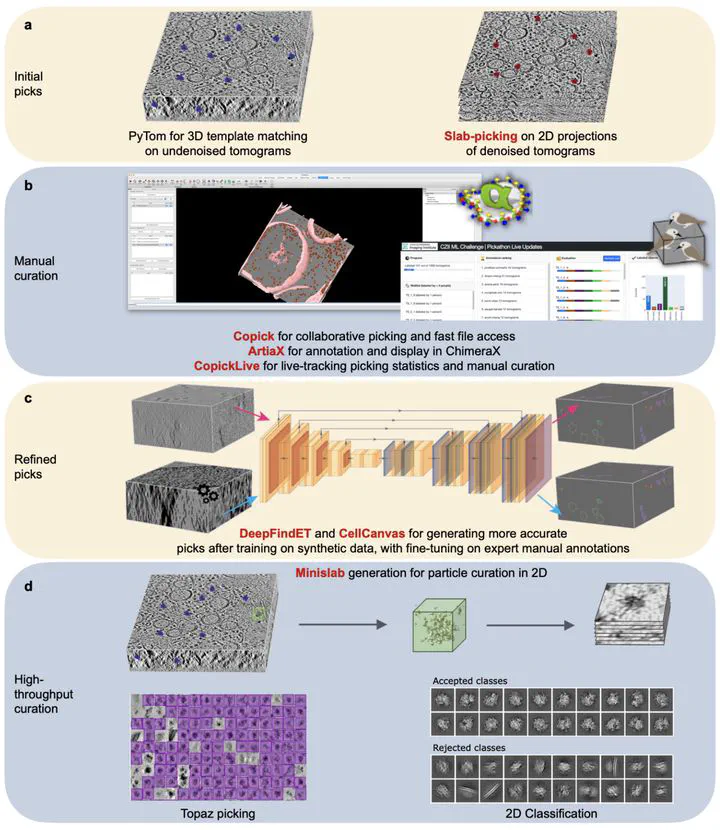 Image credit: Peck et al, 2025, Nature Methods
Image credit: Peck et al, 2025, Nature MethodsAbstract
Cryo-electron tomography (cryoET) has emerged as a powerful structural biology tool for understanding protein complexes in their native cellular environments. Presently, 3D volumes of cellular environments can be acquired in the thousands in a few days where each volume provides a rich and complex cellular landscape. Despite numerous innovations, localizing and identifying the vast majority of protein species in these volumes remains prohibitively difficult. Machine learning-based methods provide an opportunity to automate the process of labeling and annotating cryoET volumes. Due to current bottlenecks in the annotation process, and a lack of large standardized datasets, training datasets for machine learning algorithms have been scarce. Here, we present a defined “phantom” sample, along with “ground truth” annotations, that will be the basis of a machine learning challenge to bring cryoET and ML experts together and spur creativity to address this annotation problem. We have also set up a cryoET data portal that provides additional diverse sets of annotated 3D volumes from cryoET experts across the world for the machine learning challenge.
Type
Publication
Nature Methods