About Me
I’m a research scientist at Biohub building foundation models and ML infrastructure for biological imaging—focusing on 3D methods for understanding cellular structure from microscopy data.
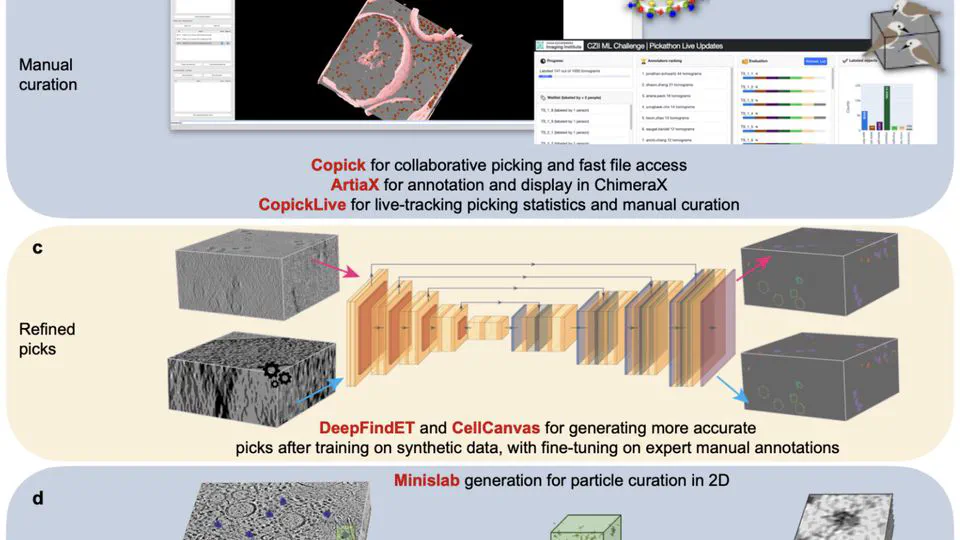
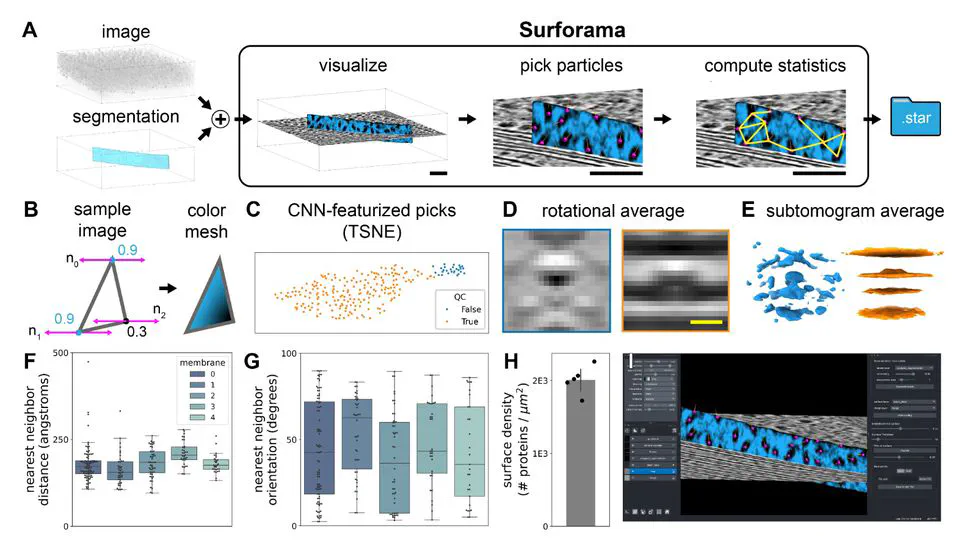
Recent work includes: Led the CZII CryoET Object Identification competition on Kaggle (published in Nature Methods, 1200+ participating teams). Built CryoLens, a generative model for learning interpretable 3D representations from cryo-electron tomography data. Created the copick ecosystem for collaborative annotation and data management in cryo-electron tomography.
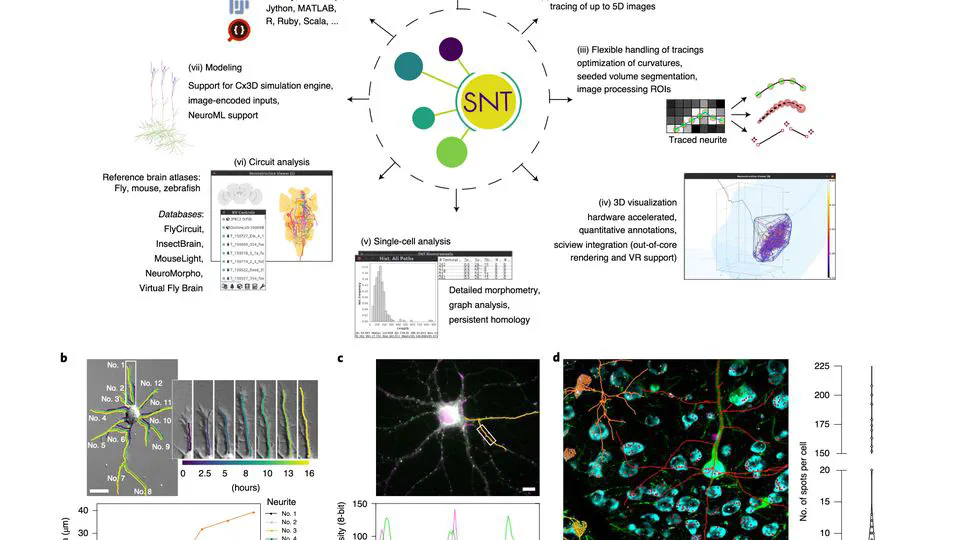
I serve on napari’s steering council and have built multiple open-source tools: sciview (3D/VR for ImageJ), SNT (neuron tracing and morphology analysis), and album (reproducible scientific workflows).
Background: Led research teams at Oak Ridge National Lab and Max Delbrück Center Berlin. Advised DARPA programs in lifelong learning and adversarial ML. PhD from Brandeis (computer science + quantitative biology), postdoc at Harvard Medical School.
- 3D and volumetric deep learning
- Foundation models for microscopy
- Open-source scientific software
- Biological image analysis
Postdoctoral Fellow, Pathology
Harvard Medical School
PhD Computer Science
Brandeis University
MA Computer Science
Brandeis University
BA
Hampshire College